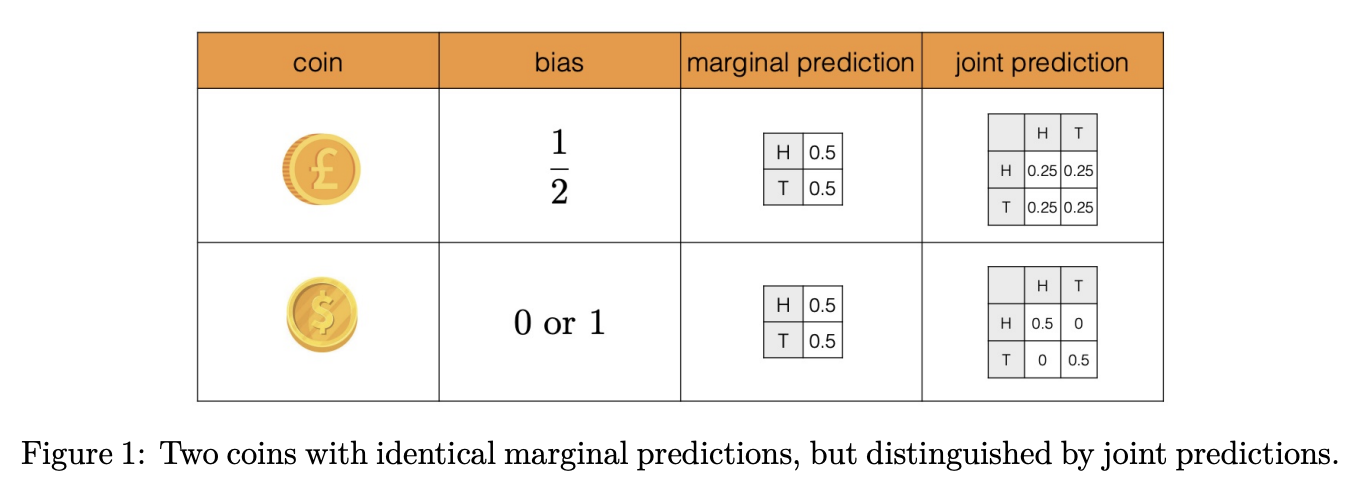
Submodularity & Batch Acquisition
(BatchBALD & Stochastic Batch Acquisition)
Why Batch Acquisition?
\[ \require{mathtools} \DeclareMathOperator{\opExpectation}{\mathbb{E}} \newcommand{\E}[2]{\opExpectation_{#1} \left [ #2 \right ]} \newcommand{\simpleE}[1]{\opExpectation_{#1}} \newcommand{\implicitE}[1]{\opExpectation \left [ #1 \right ]} \DeclareMathOperator{\opVar}{\mathrm{Var}} \newcommand{\Var}[2]{\opVar_{#1} \left [ #2 \right ]} \newcommand{\implicitVar}[1]{\opVar \left [ #1 \right ]} \newcommand\MidSymbol[1][]{% \:#1\:} \newcommand{\given}{\MidSymbol[\vert]} \DeclareMathOperator{\opmus}{\mu^*} \newcommand{\IMof}[1]{\opmus[#1]} \DeclareMathOperator{\opInformationContent}{H} \newcommand{\ICof}[1]{\opInformationContent[#1]} \newcommand{\xICof}[1]{\opInformationContent(#1)} \newcommand{\sicof}[1]{h(#1)} \DeclareMathOperator{\opEntropy}{H} \newcommand{\Hof}[1]{\opEntropy[#1]} \newcommand{\xHof}[1]{\opEntropy(#1)} \DeclareMathOperator{\opMI}{I} \newcommand{\MIof}[1]{\opMI[#1]} \DeclareMathOperator{\opTC}{TC} \newcommand{\TCof}[1]{\opTC[#1]} \newcommand{\CrossEntropy}[2]{\opEntropy(#1 \MidSymbol[\Vert] #2)} \DeclareMathOperator{\opKale}{D_\mathrm{KL}} \newcommand{\Kale}[2]{\opKale(#1 \MidSymbol[\Vert] #2)} \DeclareMathOperator{\opJSD}{D_\mathrm{JSD}} \newcommand{\JSD}[2]{\opJSD(#1 \MidSymbol[\Vert] #2)} \DeclareMathOperator{\opp}{p} \newcommand{\pof}[1]{\opp(#1)} \newcommand{\pcof}[2]{\opp_{#1}(#2)} \newcommand{\hpcof}[2]{\hat\opp_{#1}(#2)} \DeclareMathOperator{\opq}{q} \newcommand{\qof}[1]{\opq(#1)} \newcommand{\qcof}[2]{\opq_{#1}(#2)} \newcommand{\varHof}[2]{\opEntropy_{#1}[#2]} \newcommand{\xvarHof}[2]{\opEntropy_{#1}(#2)} \newcommand{\varMIof}[2]{\opMI_{#1}[#2]} \DeclareMathOperator{\opf}{f} \newcommand{\fof}[1]{\opf(#1)} \newcommand{\indep}{\perp\!\!\!\!\perp} \newcommand{\Y}{Y} \newcommand{\y}{y} \newcommand{\X}{\boldsymbol{X}} \newcommand{\x}{\boldsymbol{x}} \newcommand{\w}{\boldsymbol{\theta}} \newcommand{\W}{\boldsymbol{\Theta}} \newcommand{\wstar}{\boldsymbol{\theta^*}} \newcommand{\D}{\mathcal{D}} \newcommand{\HofHessian}[1]{\opEntropy''[#1]} \newcommand{\specialHofHessian}[2]{\opEntropy''_{#1}[#2]} \newcommand{\HofJacobian}[1]{\opEntropy'[#1]} \newcommand{\specialHofJacobian}[2]{\opEntropy'_{#1}[#2]} \newcommand{\indicator}[1]{\mathbb{1}\left[#1\right]} \]
Real-World Impact
- Labeling data is expensive ($200+/hour for medical experts)
- Need to request labels in batches for efficiency
- Naive batch selection often wastes resources
What You’ll Learn Today
After this lecture, you’ll be able to:
- Understand why naive batch acquisition fails
- Apply information theory to batch selection
- Use submodularity for efficient algorithms
- Implement BatchBALD in practice
Common Misconceptions
Warning
Batch acquisition is NOT:
- Simply selecting top-k points independently
- Only about computational efficiency
- Limited to specific acquisition functions
Batch Acquisition
Top-K Algorithm
Pseudocode
def naive_batch_acquisition(pool_set, batch_size, model):
# Score all points independently
scores = []
for x in pool_set:
score = acquisition_function(x, model)
scores.append((x, score))
# Sort by score and take top-k
scores.sort(key=lambda x: x[1], reverse=True)
selected = [x for x,_ in scores[:batch_size]]
return selectedThe Problem with Top-K
- Selects points independently
- Ignores redundancy between points
- Can waste labeling budget
- Gets worse with larger batch sizes
Redundancy Issue
Redundancy Issue
Consider MNIST digits:
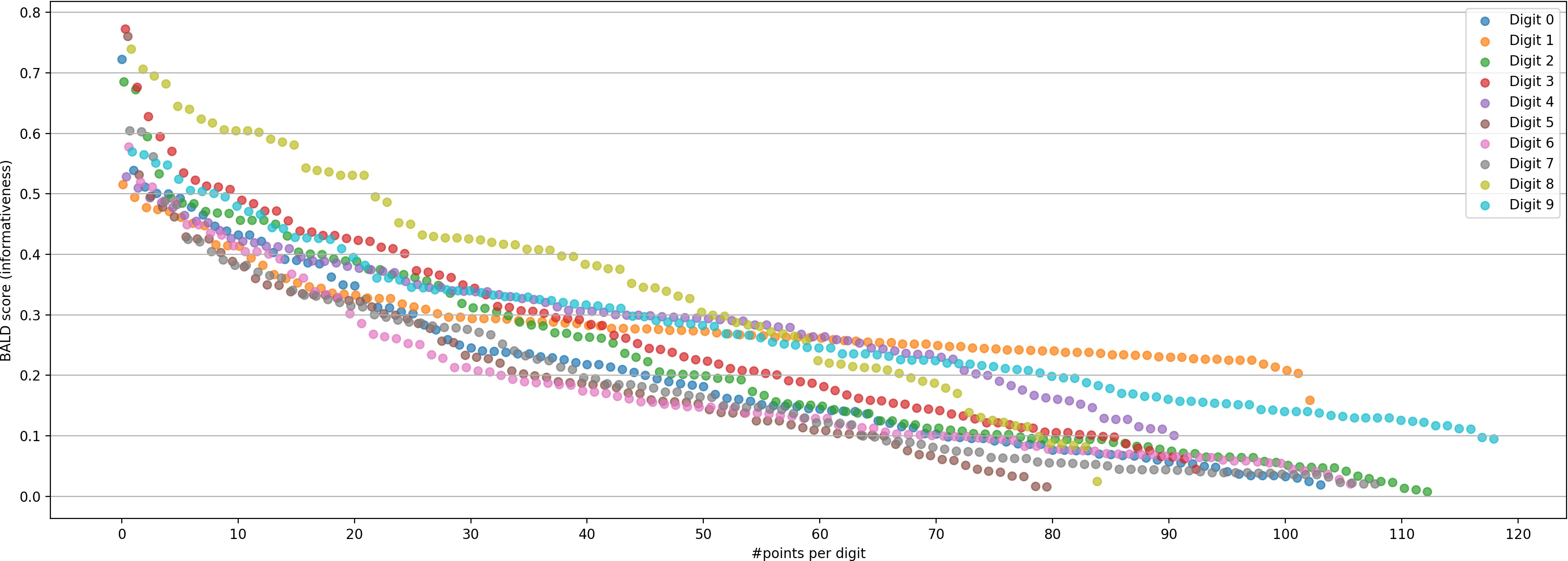
Note
- Top-K selects many “8”s
- All provide similar information overall vs other digits
- Wastes labeling budget
- Better to select diverse examples
Simple Baseline
- Bucket by predicted class
- Select examples by highest acquisition function score in each bucket round-robin.
Batch Size Effect
Small Batch Size
- Easier to contain only similar points
- More vulnerable to local redundancy
- May need more acquisition rounds
Large Batch Size
- Unlikely to contain only similar points
- Natural diversity through size
- More expensive labelling
Promise of Good Batch Acquisition
Saving Time and Labelling Cost
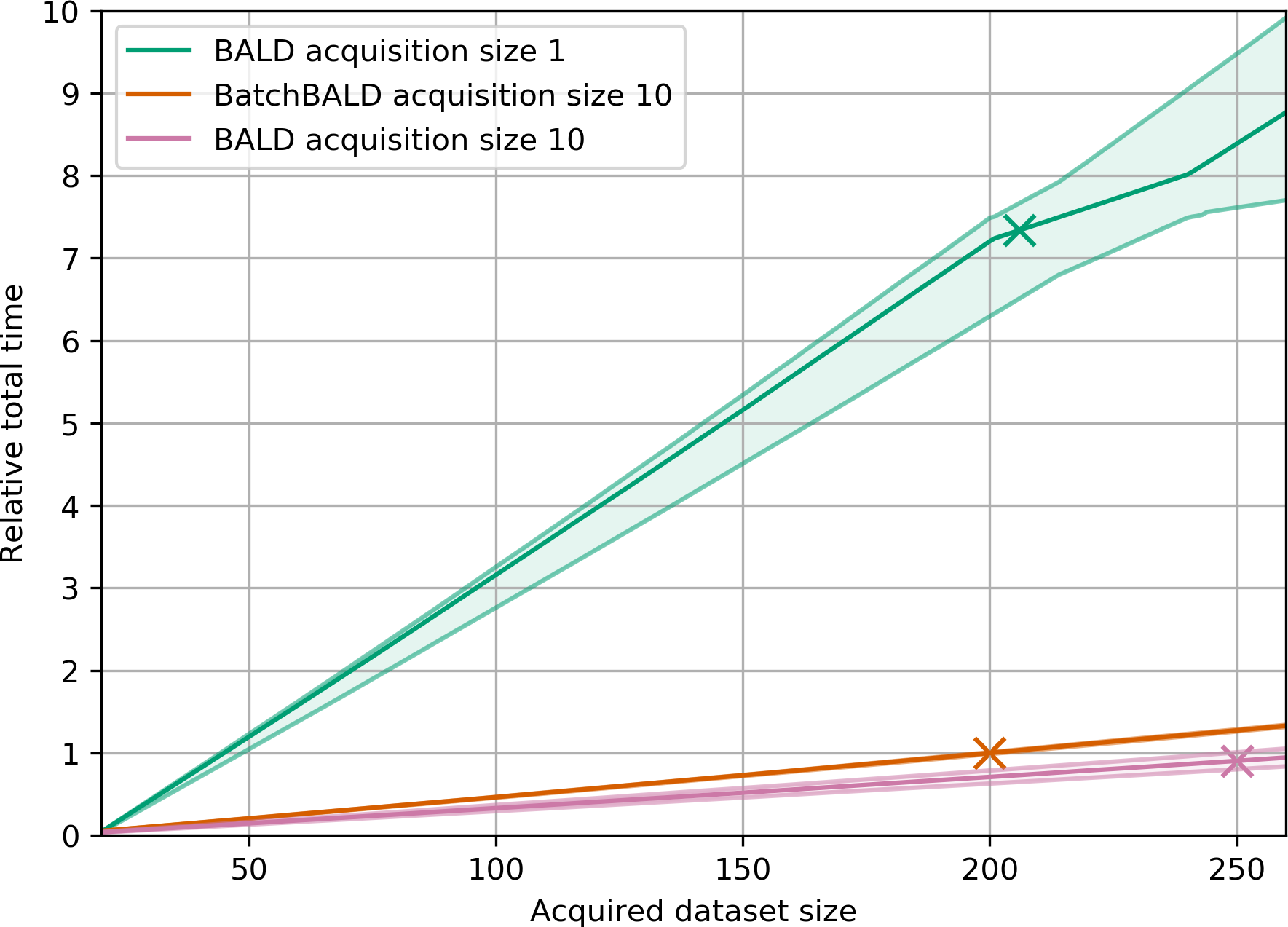
Information-Theoretic Perspective
BALD (≙ EIG)
For random variables \(\Y\) (labels) and \(\W\) (model parameters):
\[ \MIof{\Y; \W \given \x, \D} = \Hof{\Y \given \x, \D} - \E{\pof{\w \given \D}}{\Hof{\Y \given \x, \w}} \]
Note
This measures how much knowing the model parameters reduces our uncertainty about labels
Top-K Sum Equivalence
Let \(\mathcal{A}_k\) be a batch of \(k\) points.
Taking the top-k points by score is equivalent to:
\[ \arg \max_{\mathcal{A}_k \subseteq \mathcal{P}, |\mathcal{A}_k| = k} \sum_{x \in \mathcal{A}_k} \MIof{\Y; \W \given x, \D} \]
Key Insight
This shows why Top-K fails: it assumes information from each point adds independently!
Top-K Intuition
\[ \sum_{x \in \mathcal{A}_k} \MIof{\Y; \W \given x, \D} \]
Batch Intuition
How can we avoid double-counting information?
We want:
\[ \IMof{\W \cap (\cup_{x \in \mathcal{A}_k} \Y \given \x)} \]
I-Diagram \(\Rightarrow\) Information Quantity
\[ \IMof{\W \cap (\cup_{x \in \mathcal{A}_k} \Y \given \x)} \]
\[ \implies \equiv \MIof{\Y_1, .., \Y_k; \W \given \x_1, .., \x_k, \D}. \]
Joint Mutual Information
Definition 1 (BatchBALD) BatchBALD is the mutual information between the model parameters and the joint predictions of a batch of points:
\[ \MIof{\Y_1, .., \Y_k; \W \given \x_1, .., \x_k, \D} \]
Comparison
BALD
- Scores independently
- Double counts
- \(\sum_{i} \MIof{\Y_i; \W \given \x_i, \D}\)
BatchBALD
- Scores jointly
- Accounts for redundancy
- \(\MIof{\Y_1, .., \Y_k; \W \given \x_1, .., \x_k, \D}\)
Acquisition Function
\[\arg \max_{\mathcal{A}_k \subseteq \mathcal{P}, |\mathcal{A}_k| = k} \MIof{\Y_1, .., \Y_k; \W \given \x_1, .., \x_k, \D}\]
Problem: combinatorial search over all subsets!
🙀😱
What’s a cheap way to find a batch?
Greedy Optimization
def greedy_batch_acquisition(pool_set, k, acquisition_fn):
batch = []
remaining = set(pool_set)
for _ in range(k):
# Find point that maximizes marginal gain
best_x = max(remaining,
key=lambda x: acquisition_fn(batch + [x]))
batch.append(best_x)
remaining.remove(best_x)
return batchKey Properties
- Simple to implement
- Works well in practice
Guarantees
It actually works well in practice and achieves approximately a \[ 1-\frac{1}{e} \approx 0.63 \] approximation ratio of the optimal batch!
Submodularity
Greedy optimization of submodular functions yields a \[ 1-\frac{1}{e} \approx 0.63 \] approximation of the optimum!
What is Submodularity?
Submodularity captures “diminishing returns”:
Adding elements to larger sets gives less benefit
Submodularity Visualization
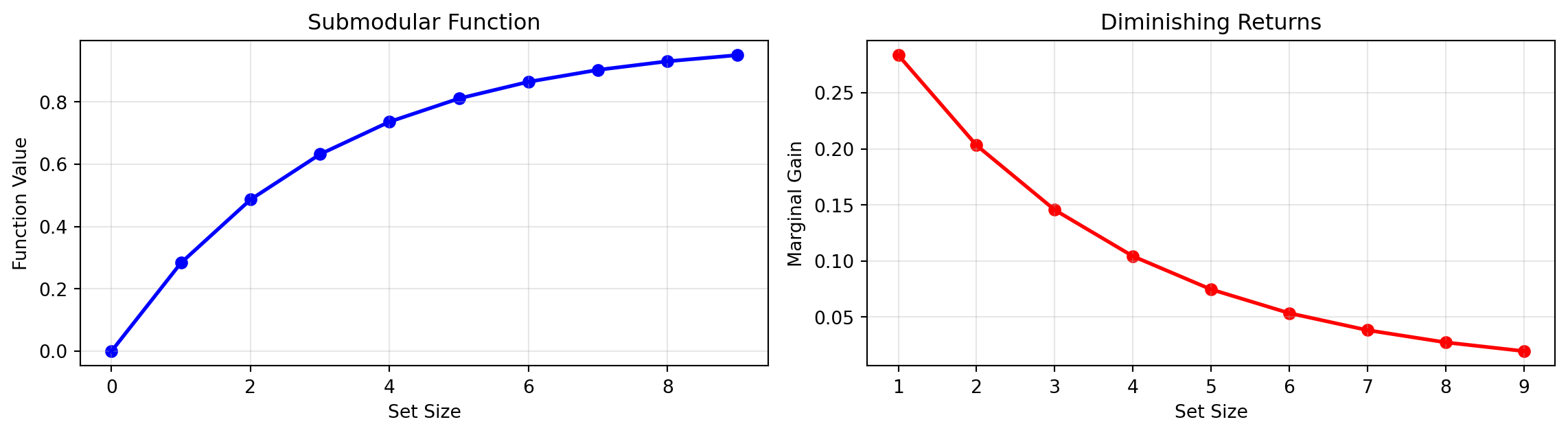
Left: Total value increases sublinearly
Right: Marginal gain decreases with set size
Formal Definition
Nemhauser, Wolsey, and Fisher (1978):
Definition 2 A set function \(f\) is submodular when \(\forall A, B\):
\[ f(A \cup B) + f(A \cap B) \leq f(A) + f(B). \]
More Intuitive Definition?
Intuitively:
- Adding elements to larger sets gives fewer benefits
- For \(X\) and \(Y_1 \subseteq Y_2\), such that \(X\) and \(Y_2\) are disjoint (\(X \cap Y_2 = \emptyset\)):
- “\(f(X \given Y_2) \leq f(X \given Y_1)\).”
- Formal: \[ f(X \cup Y_2) - f(Y_2) \leq f(X \cup Y_1) - f(Y_1). \]
Formal More Intuitive Definition
Theorem 1 For \(X\) and \(Y_1 \subseteq Y_2\), such that \(X\) and \(Y_2\) are disjoint (\(X \cap Y_2 = \emptyset\)), \(f\) is submodular iff:
\[ f(X \cup Y_2) - f(Y_2) \leq f(X \cup Y_1) - f(Y_1). \]
If we define the marginal gain of adding \(X\) to \(Y\):
\[ \Delta(X \given Y) \coloneqq f(X \cup Y) - f(Y), \]
then this is equivalent to:
\[ \Delta(X \given Y_1) \leq \Delta(X \given Y_2). \]
Proof of Equivalence
Proof (Formal Definition \(\implies\) Intuitive Definition).
Assume \(f\) is submodular as per the formal definition:
\[ f(A \cup B) + f(A \cap B) \leq f(A) + f(B). \]
For \(X\) and \(Y_1 \subseteq Y_2\), such that \(X\) and \(Y_2\) are disjoint (\(X \cap Y_2 = \emptyset\)), let:
- \(A \coloneqq X \cup Y_1\)
- \(B \coloneqq Y_2\)
Given that \(Y_1 \subseteq Y_2\) and \(X \cap Y_2 = \emptyset\), we have:
- \(A \cup B = X \cup Y_1 \cup Y_2 = X \cup Y_2\)
- \(A \cap B = (X \cup Y_1) \cap Y_2 = (X \cap Y_2) \cup (Y_1 \cap Y_2) = Y_1\)
Starting from the formal submodular inequality:
\[ \begin{aligned} f(A \cup B) + f(A \cap B) &\leq f(A) + f(B) \\ \iff f(X \cup Y_2) + f(Y_1) &\leq f(X \cup Y_1) + f(Y_2) \\ \iff f(X \cup Y_2) - f(Y_2) &\leq f(X \cup Y_1) - f(Y_1). \end{aligned} \]
This matches the intuitive definition. \(\square\)
Proof (Intuitive Definition \(\implies\) Formal Definition).
Assume \(f\) satisfies the intuitive definition:
\[ f(X \cup Y_2) - f(Y_2) \leq f(X \cup Y_1) - f(Y_1), \]
for all \(X\) and \(Y_1 \subseteq Y_2\), such that \(X\) and \(Y_2\) are disjoint (\(X \cap Y_2 = \emptyset\)).
Let’s consider arbitrary sets \(A\) and \(B\). Define:
- \(X \coloneqq A \setminus B\)
- \(Y_1 \coloneqq A \cap B\)
- \(Y_2 \coloneqq B\)
Now compute:
- \(X \cup Y_1 = (A \setminus B) \cup (A \cap B) = A\)
- \(X \cup Y_2 = (A \setminus B) \cup B = A \cup B\)
Observe that:
- \(X \cap Y_2 = (A \setminus B) \cap B = \emptyset\)
- \(Y_1 \subseteq Y_2\)
Thus, the intuitive definition implies:
\[ \begin{aligned} f(X \cup Y_2) - f(Y_2) &\leq f(X \cup Y_1) - f(Y_1) \\ \iff f(A \cup B) - f(B) &\leq f(A) - f(A \cap B) \\ \iff f(A \cup B) + f(A \cap B) &\leq f(A) + f(B). \end{aligned} \]
This matches the formal definition. \(\square\)
Simpler Definition
But wait, there is more—we can simplify this further:
Theorem 2 For any \(Y_1 \subseteq Y_2\) and \(e \notin Y_2\), \(f\) is submodular iff:
\[ f(Y_2 \cup \{e\}) - f(Y_2) \leq f(Y_1 \cup \{e\}) - f(Y_1). \]
That is:
\[ \Delta(e \given X) \leq \Delta(e \given Y). \]
Proof
\(\implies\) follows immediately from the previous definition.
\(\impliedby\):
We assume that for any \(Y_1 \subseteq Y_2\) and \(e \notin Y_2\):
\[ \Delta(e \given Y_2) \leq \Delta(e \given Y_1). \]
For \(X\) and \(Y_1 \subseteq Y_2\), such that \(X\) and \(Y_2\) are disjoint (\(X \cap Y_2 = \emptyset\)), let:
\[ \begin{aligned} S &\coloneqq Y_2 \setminus Y_1, \\ S &\coloneqq \{s_1, \ldots, s_n\}, \\ S_k &\coloneqq \{s_1, \ldots, s_k\} \subseteq S. \end{aligned} \]
For all \(k\), we have:
\[ \begin{aligned} \Delta(s_k \given X \cup Y_1 \cup S_{k-1}) &\leq \Delta(s_k \given Y_1 \cup S_{k-1}), \end{aligned} \]
as \(Y_1 \cup S_{k-1} \subseteq X \cup Y_1 \cup S_{k-1}\).
- Telescoping
- Telescoping is a technique where terms in a sequence cancel out except for the first and last terms. The name comes from how a telescope’s segments collapse into each other.
Using telescoping, we get:
\[ \begin{aligned} \sum_{k=1}^{n} \Delta(s_k \given X \cup Y_1 \cup S_{k-1}) &\leq \sum_{k=1}^{n} \Delta(s_k \given Y_1 \cup S_{k-1}) \\ \iff \sum_{k=1}^{n} f(X \cup Y_1 \cup S_{k-1} \cup \{s_k\}) &\\ - f(X \cup Y_1 \cup S_{k-1}) &\leq \sum_{k=1}^{n} f(Y_1 \cup S_{k-1} \cup \{s_k\}) \\ &\hphantom{\leq} - f(Y_1 \cup S_{k-1}) \\ \iff \sum_{k=1}^{n} f(X \cup Y_1 \cup S_{k}) &\\ - f(X \cup Y_1 \cup S_{k-1}) &\leq \sum_{k=1}^{n} f(Y_1 \cup S_{k}) \\ & \hphantom{\leq} - f(Y_1 \cup S_{k-1}) \\ \iff f(X \cup Y_1 \cup S) &\\ - f(X \cup Y_1) &\leq f(Y_1 \cup S) - f(Y_1) \\ \iff f(X \cup Y_2) &\\ - f(X \cup Y_1) &\leq f(Y_2) - f(Y_1), \end{aligned} \]
where we have used that \(Y_2 = Y_1 \cup S \leftrightarrow S = Y_2 \setminus Y_1\).
Rearranging, we get:
\[ f(X \cup Y_2) - f(Y_2) \leq f(X \cup Y_1) - f(Y_1). \]
\(\square\)
Summary
This characterizes submodularity in terms of marginal gains:
\[ \begin{aligned} f &\text{ is submodular} \\ \iff f(A \cup B) + f(A \cap B) &\leq f(A) + f(B) \\ \iff \Delta(X \given Y_2) &\leq \Delta(X \given Y_1) \\ \iff \Delta(e \given Y_2) &\leq \Delta(e \given Y_1). \end{aligned} \]
for \(Y_1 \subseteq Y_2\) and \(e \notin Y_2\) and \(X \cap Y_2 = \emptyset\).
\(1 - \frac{1}{e}\) Optimality
Theorem 3 For a monotone submodular function \(f\) with \(f(\emptyset) = 0\), the greedy algorithm achieves at least:
\[ f(\mathcal{A}_k^{\text{greedy}}) \geq \left(1 - \frac{1}{e}\right) f(\mathcal{A}_k^*) \]
where \(\mathcal{A}_k^*\) is the optimal set of size \(k\).
Key Insights
- Greedy selection achieves ≈63% of optimal value
- Holds even for very large search spaces
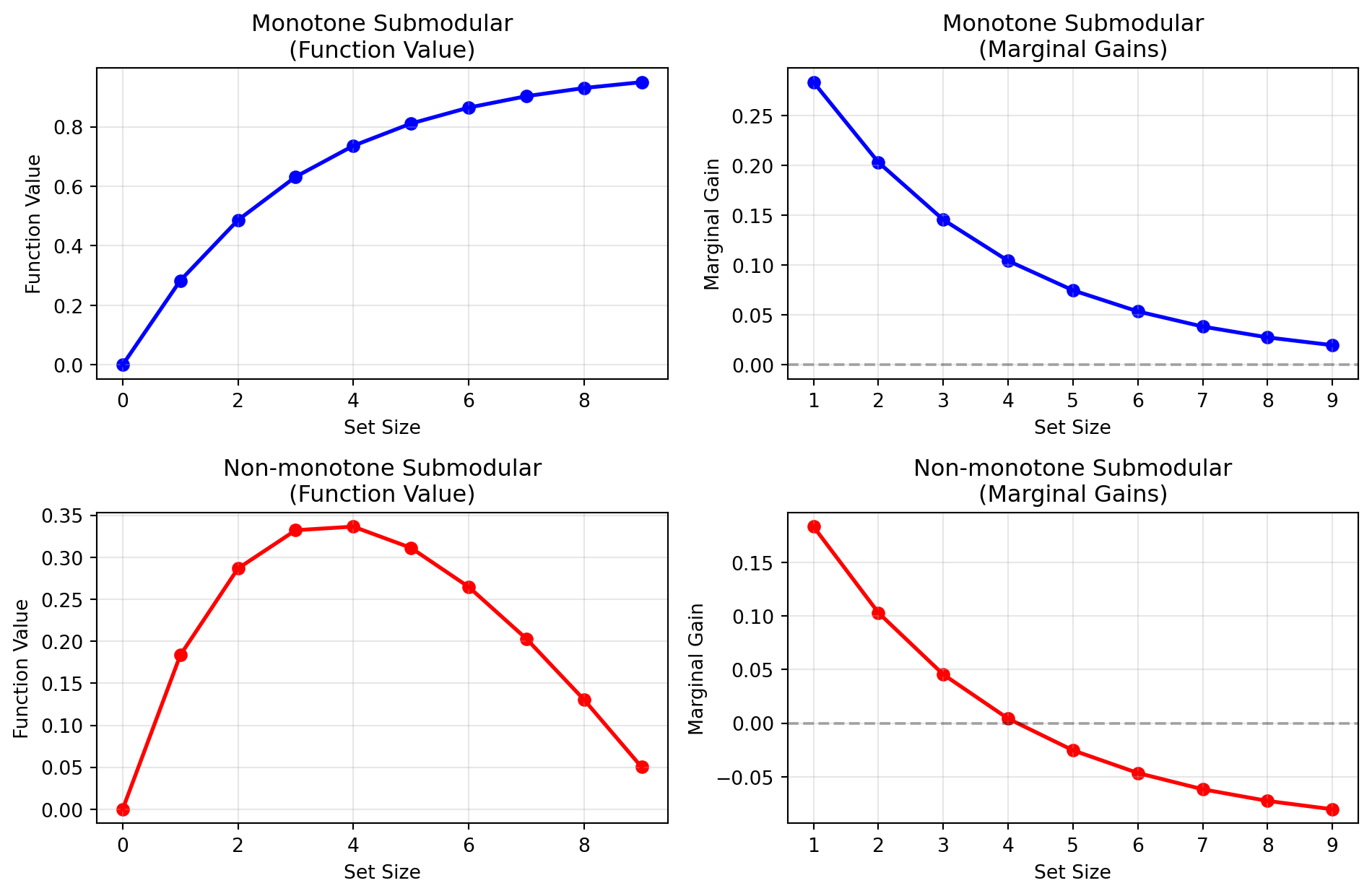
Monotone Submodular Functions
Definition 3 A set function \(f: 2^V \rightarrow \mathbb{R}\) is monotone if for all \(A \subseteq B \subseteq V\):
\[ f(A) \leq f(B) \]
Intuition
- Adding elements never hurts \(\leftrightarrow\) non-negative gains!
- Function value can only increase
- Different from diminishing returns!
Common Confusion
Submodularity: diminishing returns
Monotonicity: always increasing
Both needed for greedy guarantees!
Questions
- What does a non-monotone submodular function look like?
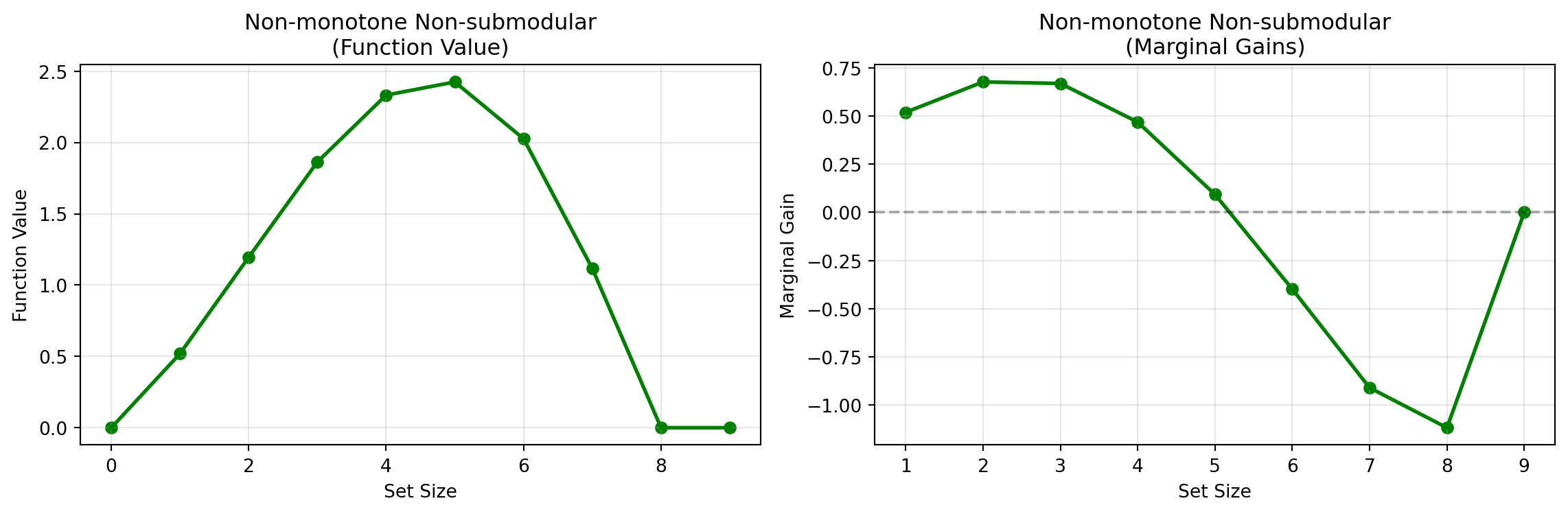
- What does a non-monotone non-submodular function look like?
Submodular Non-/Monotone
Non-Submodular Non-Monotone
Optimality Proof
Statement
Let \(f: 2^{\mathcal{V}} \rightarrow \mathbb{R}_{\geq 0}\) be a monotone (i.e., \(f(A) \leq f(B)\) whenever \(A \subseteq B\)), and submodular function with \(f(\emptyset) = 0\) defined over a finite ground set \(\mathcal{V}\).
Our goal is to select a subset \(S \subseteq \mathcal{V}\) of size \(k\) that maximizes \(f(S)\):
\[ S^* = \arg\max_{|S| \leq k} f(S). \]
Greedy Algorithm
The greedy algorithm constructs a set \(S_{\text{greedy}}\) by starting with \(S_0 = \emptyset\) and iteratively adding elements:
\[ S_{i} = S_{i-1} \cup \{ e_i \}, \]
where \(e_i\) is chosen to maximize the marginal gain:
\[ \begin{aligned} e_i &\coloneqq \arg\max_{e \in \mathcal{V} \setminus S_{i-1}} \left[ f(S_{i-1} \cup \{ e \}) - f(S_{i-1}) \right] \\ &= \arg\max_{e \in \mathcal{V} \setminus S_{i-1}} \Delta(e \mid S_{i-1}). \end{aligned} \]
Goal
We will show that:
\[ f(S_{\text{greedy}}) \geq \left( 1 - \frac{1}{e} \right) f(S^*). \]
Key Concepts
Marginal Gain: The benefit of adding an element \(e\) to a set \(A\):
\[ \Delta(e \mid A) \coloneqq f(A \cup \{ e \}) - f(A). \]
Submodularity: Diminishing returns property, i.e., for \(A \subseteq B\), we have:
\[ \Delta(e \mid A) \geq \Delta(e \mid B). \]
Proof Sketch
Bounding the Marginal Gain
At each iteration \(i\):
\[ \Delta(e_i \mid S_{i-1}) \geq \frac{1}{k} \left[ f(S^*) - f(S_{i-1}) \right]. \]
Recursive Inequality
Update the function value:
\[ f(S_i) \geq f(S_{i-1}) + \frac{1}{k} \left[ f(S^*) - f(S_{i-1}) \right]. \]
Unfolding the Recursion
After \(k\) iterations:
\[ f(S_{\text{greedy}}) \geq \left( 1 - \left( 1 - \frac{1}{k} \right)^k \right) f(S^*). \]
Limit Evaluation
As \(k\) increases:
\[ \left( 1 - \frac{1}{k} \right)^k \leq \frac{1}{e}, \]
so:
\[ f(S_{\text{greedy}}) \geq \left( 1 - \frac{1}{e} \right) f(S^*). \]
Proof of \(\left(1 - \frac{1}{e}\right)\)-Optimality
Step 1: Bounding the Marginal Gain
At iteration \(i\), consider the optimal set \(S^*\) and the current greedy set \(S_{i-1}\).
Define \(R \coloneqq S^* \setminus S_{i-1}\).
Since \(|S^*| = k\), we have \(|R| \leq k\).
By submodularity and the fact that \(f\) is monotone:
\[ \begin{align*} f(S^*) - f(S_{i-1}) &\leq f(S_{i-1} \cup R) - f(S_{i-1}) \\ &\leq \sum_{e \in R} \Delta(e \mid S_{i-1}). \end{align*} \tag{1}\]
Explanation:
First inequality from monotonicity.
Second inequality from submodularity after telescoping:
\[ \begin{aligned} &f(S_{i-1} \cup R) - f(S_{i-1}) \\ &\quad = \sum_{i=1}^{|R|} f(S_{i-1} \cup \{r_1, \ldots, r_i\}) \\ &\quad \phantom{= \sum} - f(S_{i-1} \cup \{r_1, \ldots, r_{i-1}\}) \\ &\quad = \sum_{i=1}^{|R|} \Delta(r_i \mid S_{i-1} \cup \{r_1, \ldots, r_{i-1}\}) \\ &\quad \leq \sum_{i=1}^{|R|} \Delta(r_i \mid S_{i-1})\\ &\quad = \sum_{e \in R} \Delta(e \mid S_{i-1}). \end{aligned} \]
Since the greedy choice \(e_i\) maximizes \(\Delta(e \mid S_{i-1})\):
\[ \begin{aligned} \Delta(e_i \mid S_{i-1}) &\geq \frac{1}{|R|} \sum_{e \in R} \Delta(e \mid S_{i-1}) \\ &\geq \frac{1}{k} \left[ f(S^*) - f(S_{i-1}) \right]. \end{aligned} \]
Explanation:
First inequality from maximality:
\[ \Delta(e_i \mid S_{i-1}) \geq \Delta(e \mid S_{i-1}), \]
and summing over all \(e \in R\):
\[ |R| \Delta(e_i \mid S_{i-1}) \geq \sum_{e \in R} \Delta(e \mid S_{i-1}). \]
Second inequality follows from \(|R| \leq k\) and Equation 1 above.
Step 2: Recursive Inequality
From the above bound:
\[ \begin{align*} f(S_i) &= f(S_{i-1}) + \Delta(e_i \mid S_{i-1}) \\ &\geq f(S_{i-1}) + \frac{1}{k} \left[ f(S^*) - f(S_{i-1}) \right] \\ &= \left( 1 - \frac{1}{k} \right) f(S_{i-1}) + \frac{1}{k} f(S^*). \end{align*} \]
Step 3: Unfolding the Recursion
Define \(\gamma_i \coloneqq f(S^*) - f(S_i)\).
Then:
\[ \gamma_i \leq \left( 1 - \frac{1}{k} \right) \gamma_{i-1}. \]
Note
Explanation:
We subtract the previous inequality
\[ f(S_i) \geq \left( 1 - \frac{1}{k} \right) f(S_{i-1}) + \frac{1}{k} f(S^*) \]
from \(f(S^*)\)
which yields:
\[ \begin{aligned} &f(S^*) - f(S_i) \\ &\quad \leq f(S^*) - \left[ \left( 1 - \frac{1}{k} \right) f(S_{i-1}) + \frac{1}{k} f(S^*) \right] \\ &\quad = \left( 1 - \frac{1}{k} \right) \left[ f(S^*) - f(S_{i-1}) \right]. \end{aligned} \]
which is just the definition of \(\gamma_i\) on the LHS and \(\gamma_{i-1}\) on the RHS:
\[ \gamma_i \leq \left( 1 - \frac{1}{k} \right) \gamma_{i-1}. \]
By iterating this inequality:
\[ \gamma_k \leq \left( 1 - \frac{1}{k} \right)^k f(S^*). \]
Explanation:
We start with
\[ \gamma_0 = f(S^*) - f(S_0) = f(S^*), \]
as \(f(S_0) = f(\emptyset) = 0\).
Then we apply the inequality \(\gamma_i \leq \left( 1 - \frac{1}{k} \right) \gamma_{i-1}\) iteratively to obtain:
\[ \begin{aligned} \gamma_k &\leq \left( 1 - \frac{1}{k} \right) \left( 1 - \frac{1}{k} \right) \cdots \left( 1 - \frac{1}{k} \right) \gamma_0 \\ &= \left( 1 - \frac{1}{k} \right)^k \gamma_0. \end{aligned} \]
Thus, the greedy solution satisfies:
\[ \begin{aligned} f(S_{\text{greedy}}) &= f(S_k) \\ &= f(S^*) - \gamma_k \\ &\geq f(S^*) \left( 1 - \left( 1 - \frac{1}{k} \right)^k \right). \end{aligned} \]
Explanation:
We start with \(\gamma_k = f(S^*) - f(S_k)\).
Then we apply the itereated inequality to obtain:
\[ \begin{aligned} f(S^*) - \gamma_k &\geq f(S^*) - \left( 1 - \frac{1}{k} \right)^k f(S^*) \\ &= f(S^*) \left( 1 - \left( 1 - \frac{1}{k} \right)^k \right). \end{aligned} \]
Step 4: Limit Evaluation
Remembering that:
\[ \lim_{k \rightarrow \infty} \left( 1 - \frac{1}{k} \right)^k = \frac{1}{e}, \]
and that:
\[ \left( 1 - \frac{1}{k} \right)^k < \frac{1}{e}, \]
we conclude:
\[ \begin{aligned} f(S_{\text{greedy}}) &\geq \left( 1 - \left( 1 - \frac{1}{k} \right)^k \right) f(S^*) \\ &\geq \left( 1 - \frac{1}{e} \right) f(S^*). \end{aligned} \] \(\square\)
Summary
The greedy algorithm provides a \(\left( 1 - \frac{1}{e} \right)\)-approximation to the optimal solution when maximizing a non-negative, monotone, submodular function under a cardinality constraint.
Why?
This result is significant because it guarantees that the greedy approach to batch acquisition in settings like active learning will achieve near-optimal utility, ensuring efficient use of resources.
Submodular BatchBALD
Set
\[ f({\x_1, \ldots, \x_k}) \coloneqq \MIof{\Y_1, \ldots, \Y_k; \W \given \x_1, \ldots, \x_k}. \]
Show:
- \(f(\emptyset) = 0\)
- Monotone
- Submodular
Marginal Gain
For this definition of \(f\), the marginal gain is:
\[ \begin{aligned} \Delta(x_e \mid \x_1, .., \x_k) &= f(\{ \x_1, .., \x_k, \x_e \}) - f(\{ \x_1, .., \x_k \}) \\ &= \MIof{\Y_e, \Y_1, .., \Y_k; \W \given \x_e, \x_1, .., \x_k} \\ &\quad - \MIof{\Y_1, .., \Y_k; \W \given \x_1, .., \x_k} \\ &= \MIof{\Y_e; \W \given \x_e, \Y_1, \x_1, .., \Y_k, \x_k} \\ \end{aligned} \]
Proof (Last Step)
Convince yourself that:
\[ \MIof{A, B; \W \given C} = \MIof{A; \W \given C} + \MIof{B; \W \given A, C}. \]
And then:
\[ \MIof{B; \W \given A, C} = \MIof{A, B; \W \given C} - \MIof{A; \W \given C}. \] \(\square\)
Proof: \(f(\emptyset) = 0\)
\[ f(\emptyset) = \MIof{ \emptyset; \W} = 0. \] \(\square\)
Proof: Monotone
Show that for \(A \subseteq B\):
\[ f(A) \leq f(B). \]
Proof.
For \(x_1, .., x_n \in \mathcal{P}\) and \(1 \le k \le n, k \in \mathbb{N}\):
\[ \begin{aligned} &f(\{ \x_1, .., \x_k \}) \\ &\quad \leq f(\{ \x_1, .., \x_n \}) \\ \iff &\MIof{\Y_1, .., \Y_k; \W \given \x_1, .., \x_k} \\ &\quad \leq \MIof{\Y_1, .., \Y_{n}; \W \given \x_1, .., \x_{n}} \\ \iff &\Hof{\W} - \Hof{\W \given \Y_1, .., \Y_k, \x_1, .., \x_k} \\ &\quad \leq \Hof{\W} - \Hof{\W \given \Y_1, .., \Y_{n}, \x_1, .., \x_{n}} \\ \iff &\Hof{\W \given \Y_1, .., \Y_k, \x_1, .., \x_k} \\ &\quad \geq \Hof{\W \given \Y_1, .., \Y_{n}, \x_1, .., \x_{n}}. \end{aligned} \]
Conditioning reduces entropy.
Thus, \(f\) is monotone. \(\square\)
Alternative:
We notice that
\[ \begin{aligned} &f(\{ \x_1, .., \x_k \}) \leq f(\{ \x_1, .., \x_n \}) \\ \iff &0 \leq \Delta(\x_{k+1}, .., \x_n \mid \x_1, .., \x_k) \\ \iff &0 \leq \MIof{\Y_{k+1}, .., \Y_n; \W \given \x_{k+1}, .., \x_n, \Y_1, .., \Y_k, \x_1, .., \x_k}, \end{aligned} \]
which is true as pairwise mutual information is non-negative. \(\square\)
Proof: Submodular
For \(X\) and \(Y_1 \subseteq Y_2\) with \(X \cap Y_2 = \emptyset\), we need to show:
\[ \Delta(X \given Y_2) \leq \Delta(X \given Y_1). \]
Notation: We rename these to \(A\) and \(B_1 \subseteq B_2\) to avoid confusing with the random variables \(\X\) and \(\Y\). We drop the conditioning on \(\x\) and only write the \(\Y\) to save space.
We need to show:
\[ \MIof{\Y_{A} ; \W \given \Y_{B_2}} \leq \MIof{\Y_{A} ; \W \given \Y_{B_1}}. \]
Let:
\[ S \coloneqq B_2 \setminus B_1. \]
We can rewrite this as:
\[ \begin{aligned} &\MIof{\Y_{A} ; \W \given \Y_{B_1}} - \MIof{\Y_{A} ; \W \given \Y_{B_2}} \geq 0 \\ \iff &\MIof{\Y_{A} ; \W \given \Y_{B_1}} - \MIof{\Y_{A} ; \W \given \Y_{B_1}, \Y_{S}} \geq 0 \\ \iff &\MIof{\Y_{A} ; \W ; \Y_S \given \Y_{B_1}} \geq 0. \end{aligned} \]
Using the symmetry of the mutual information:
\[ \begin{aligned} 0 &\leq \MIof{\Y_{A} ; \W ; \Y_S \given \Y_{B_1}} \\ &= \MIof{\Y_{A} ; \Y_S; \W \given \Y_{B_1}} \\ &= \MIof{\Y_{A} ; \Y_S \given \W, \Y_{B_1}} - \MIof{\Y_{A} ; \Y_S \given \W, \Y_{B_1}} \\ &= \MIof{\Y_{A} ; \Y_S \given \W, \Y_{B_1}} \ge 0, \end{aligned} \]
because \(\MIof{\Y_{A} ; \Y_S \given \W, \Y_{B_1}} = 0\) and the pairwise mutual information is non-negative.
\(\MIof{\Y_{A} ; \Y_S \given \W, \Y_{B_1}} = 0\) because:
Recall that our probabilistic model is
\[ \pof{\y_1, .., \y_n, \w \given \x_1, .., \x_n} = \pof{\w} \prod_{i=1}^n \pof{\y_i \given \w, \x_i}. \]
Thus, for all \(i \not=j\): \[ \Y_i \indep \Y_j \given \W, \x_i, \x_j. \]
Thus, \(f\) is submodular. \(\square\)
Summary
\(f\) is submodular and monotone, that is
\[ f_{\text{BatchBALD}}({\x_1, \ldots, \x_k}) \coloneqq \MIof{\Y_1, \ldots, \Y_k; \W \given \x_1, \ldots, \x_k}. \]
Note
BatchBALD is submodular, allowing efficient computation despite the combinatorial nature of batch selection
Revisiting the Greedy Algorithm
Given \(x_1, \ldots, x_{i-1}\), at each step we find the point \(x_i\) that maximizes:
\[ f_{\text{BatchBALD}}({\x_1, \ldots, \x_{i-1}, x_i}). \]
The first \({\x_1, \ldots, \x_{i-1}}\) are already selected and thus fixed.
We can just as well maximize the marginal gain:
\[ \Delta(x \given x_1, \ldots, x_{i-1}) = \MIof{\Y; \W \given x, \Y_1, \x_1, \ldots, \Y_{i-1}, \x_{i-1}}. \]
At each step, we thus maximize the EIG conditioned on the other selected points in expectation (we don’t know the actual labels so we use the predictive distribution and take expectations wrt. the labels!).
BatchBALD in Practice
Key Components
- Joint mutual information computation
- Efficient caching strategies
- MC dropout for uncertainty
Results
Performance Gains
- More diverse batch selection
- Better data efficiency
- State-of-the-art results on standard benchmarks
Computing BatchBALD
Given some samples \(\w_1\), …, \(\w_k\), we need to compute:
\[ \MIof{\Y_1, \ldots, \Y_n; \W \given \x_1, \ldots, \x_n}. \]
Split
Because predictions are independent given the weights, we have:
\[ \begin{aligned} \MIof{\Y_1, .., \Y_n; \W \given \x_1, .., \x_n} &= \Hof{\Y_1, .., \Y_n} - \Hof{\Y_1, .., \Y_n \given \W, \x_1, .., \x_n} \\ &= \Hof{\Y_1, .., \Y_n \given \x_1, .., \x_n} - \sum_{i=1}^k \Hof{\Y_i \given \x_i, \W}. \end{aligned} \]
We can precompute \(\Hof{\Y_i \given \x_i, \W}\) for samples in the pool set and reuse these values throughout during the greedy selection process.
Computing the Joint
\(\Hof{\Y_1, .., \Y_k \given \x_1, .., \x_n}\) is more challenging:
\[ \begin{aligned} \pof{\y_1, .., \y_n \given \x_1, .., \x_n} &= \E{\pof{\w}}{\pof{\y_1, .., \y_n \given \x_1, .., \x_n, \w}} \\ &\approx \frac{1}{k} \sum_{i=1}^k \pof{\y_1, .., \y_n \given \x_1, .., \x_n, \w_i} \\ &= \frac{1}{k} \sum_{i=1}^k \prod_{j=1}^n \pof{\y_j \given \x_j, \w_i}. \end{aligned} \]
And then:
\[ \Hof{\Y_1, .., \Y_k \given \x_1, .., \x_n} = \sum_{y_1, .., y_n} -\pof{y_1, .., y_n \given \x_1, .., \x_n} \ln \pof{y_1, .., y_n \given \x_1, .., \x_n}. \]
What is the challenge with this?
MC Sampling of \(\y_1, .., \y_n\)
To reduce the complexity of the outer sum, we can sample \(\y_1, .., \y_n \sim \pof{\y_1, .., \y_n \given \x_1, .., \x_n}\) and to compute an estimate.
But this does not work well in practice. 😢
Thus,
\[ n \le 7. \]
is a common choice for BatchBALD.
(\(\implies\) stochastic sampling instead of deterministic!)
Inconsistent MC Dropout
How do we sample \(\pof{\y_1, .., \y_n \given \x_1, .., \x_n, \w_i}\)?
Dropout masks change on every call to the model ⚡️
We cannot compute the joint distributions with different masks (parameter samples) for different points:
![]()
Inconsistent parameter samples cannot capture correlations between points!
Easy Consistent Samples per Batch?
Cheap & Easy?
- could sample in larger test batches?
- would loose correlations between batches but we will
- capture correlations within a batch of points at least.⏭️
BUT:
⏭️
Consistent MC Dropout
Implement custom MC dropout that samples fixed dropout masks once when we enter model.eval() and then reuses them for all points.
We always sample multiple masks in batch to speed up sampling:
TensorType[n_samples, n_inputs, n_classes]
Pseudo-Code
class ConsistentMCDropout(nn.Module):
"""Consistent MC Dropout layer that reuses masks during evaluation."""
def __init__(self, p: float = 0.5):
super().__init__()
self.p = p
self.mask = None
self.training = True
def forward(self, x: torch.Tensor) -> torch.Tensor:
if self.training:
# Standard training behavior
return F.dropout(x, self.p, self.training)
if self.mask is None:
# Generate mask for all MC samples at once
# Shape: [n_samples, *x.shape[1:]]
self.mask = (torch.rand(x.shape, device=x.device) > self.p).float() / (1 - self.p)
return x * self.mask
class ConsistentMCModel(nn.Module):
"""Wrapper for models to enable consistent MC dropout sampling."""
def __init__(self, base_model: nn.Module):
super().__init__()
self.base_model = base_model
self.n_samples = 1
self._replace_dropouts()
@classmethod
def _module_replace_dropout(cls, module):
for name, child in module.named_children():
if isinstance(child, nn.Dropout):
setattr(module, name, ConsistentMCDropout(child.p))
else:
cls._module_replace_dropout(child)
def _replace_dropouts(self):
"""Replace all nn.Dropout with ConsistentMCDropout."""
self._module_replace_dropout(self.base_model)
def reset_masks(self):
"""Reset stored masks."""
for module in self.modules():
if isinstance(module, ConsistentMCDropout):
module.mask = None
def eval(self):
"""Set evaluation mode and prepare for MC sampling."""
super().eval()
self.reset_masks()
return self
def forward(self, x: torch.Tensor) -> torch.Tensor:
"""Forward pass with optional MC samples."""
if self.training:
return self.base_model(x)
# Expand input for MC samples
# Shape: [batch_size, ...] -> [n_samples, batch_size, ...]
x_expanded = x.unsqueeze(0).expand(self.n_samples, *x.shape)
# Get predictions for all samples
with torch.no_grad():
outputs = self.base_model(x_expanded)
return outputs # Shape: [n_samples, batch_size, n_classes]
# Usage example:
def get_consistent_mc_predictions(
model: ConsistentMCModel,
x: torch.Tensor,
n_samples: int = 100
) -> torch.Tensor:
"""Get consistent MC dropout predictions."""
model.n_samples = n_samples
model.eval()
with torch.no_grad():
predictions = model(x) # [n_samples, batch_size, n_classes]
return predictionsEfficient(?) Joint Entropy
def compute_batch_joint_entropy_matmul(
fixed_log_probs: torch.Tensor, # [n_samples, B, n_classes]
pool_log_probs: torch.Tensor, # [n_samples, pool_size, n_classes]
n_classes: int = 10
) -> torch.Tensor:
"""Compute joint entropy using matrix multiplication.
Args:
fixed_log_probs: Predictions for fixed batch [n_samples, B, n_classes]
pool_log_probs: Predictions for pool points [n_samples, pool_size, n_classes]
n_classes: Number of classes
Returns:
Joint entropies [pool_size]
"""
n_samples = fixed_log_probs.shape[0]
batch_size = fixed_log_probs.shape[1]
pool_size = pool_log_probs.shape[1]
assert fixed_log_probs.shape[0] == pool_log_probs.shape[0]
assert fixed_log_probs.shape[2] == pool_log_probs.shape[2]
assert fixed_log_probs.device == pool_log_probs.device
# Convert pool logits to probabilities
pool_probs = pool_log_probs.exp() # [n_samples, pool_size, n_classes]
# Generate combinations using meshgrid
combinations = torch.stack(
torch.meshgrid(*[torch.arange(n_classes) for _ in range(batch_size)]),
dim=-1
).reshape(-1, batch_size) # [n_classes^B, B]
# Gather probabilities for fixed batch
# Shape: [n_samples, n_classes^B, B]
gathered_preds = fixed_log_probs[:, None, :, :].expand(
n_samples, len(combinations), batch_size, n_classes
).gather(-1, combinations[None, :, :, None].expand(n_samples, -1, -1, 1)).squeeze(-1)
# Multiply probabilities along B dimension
fixed_joint_probs = gathered_preds.sum(dim=-1).exp() # [n_samples, n_classes^B]
# Process pool points in chunks
# TODO: Adjust based on available memory
chunk_size = 100
joint_entropies = torch.zeros(pool_size, device=fixed_log_probs.device)
for chunk_start in range(0, pool_size, chunk_size):
chunk_end = min(chunk_start + chunk_size, pool_size)
chunk_probs = pool_probs[:, chunk_start:chunk_end] # [n_samples, chunk_size, n_classes]
# Reshape for matrix multiplication
# [n_samples, chunk_size, n_classes] -> [chunk_size, n_classes, n_samples]
chunk_probs = chunk_probs.permute(1, 2, 0)
# Matrix multiplication and normalize
# [chunk_size, n_classes, n_samples] @ [n_samples, n_classes^B]
# = [chunk_size, n_classes, n_classes^B]
joint_probs = (chunk_probs @ fixed_joint_probs) / n_samples
# Compute entropy
# Shape: [chunk_size]
chunk_entropies = -torch.sum(
joint_probs * torch.log(joint_probs + 1e-10),
dim=(1, 2)
)
joint_entropies[chunk_start:chunk_end] = chunk_entropies
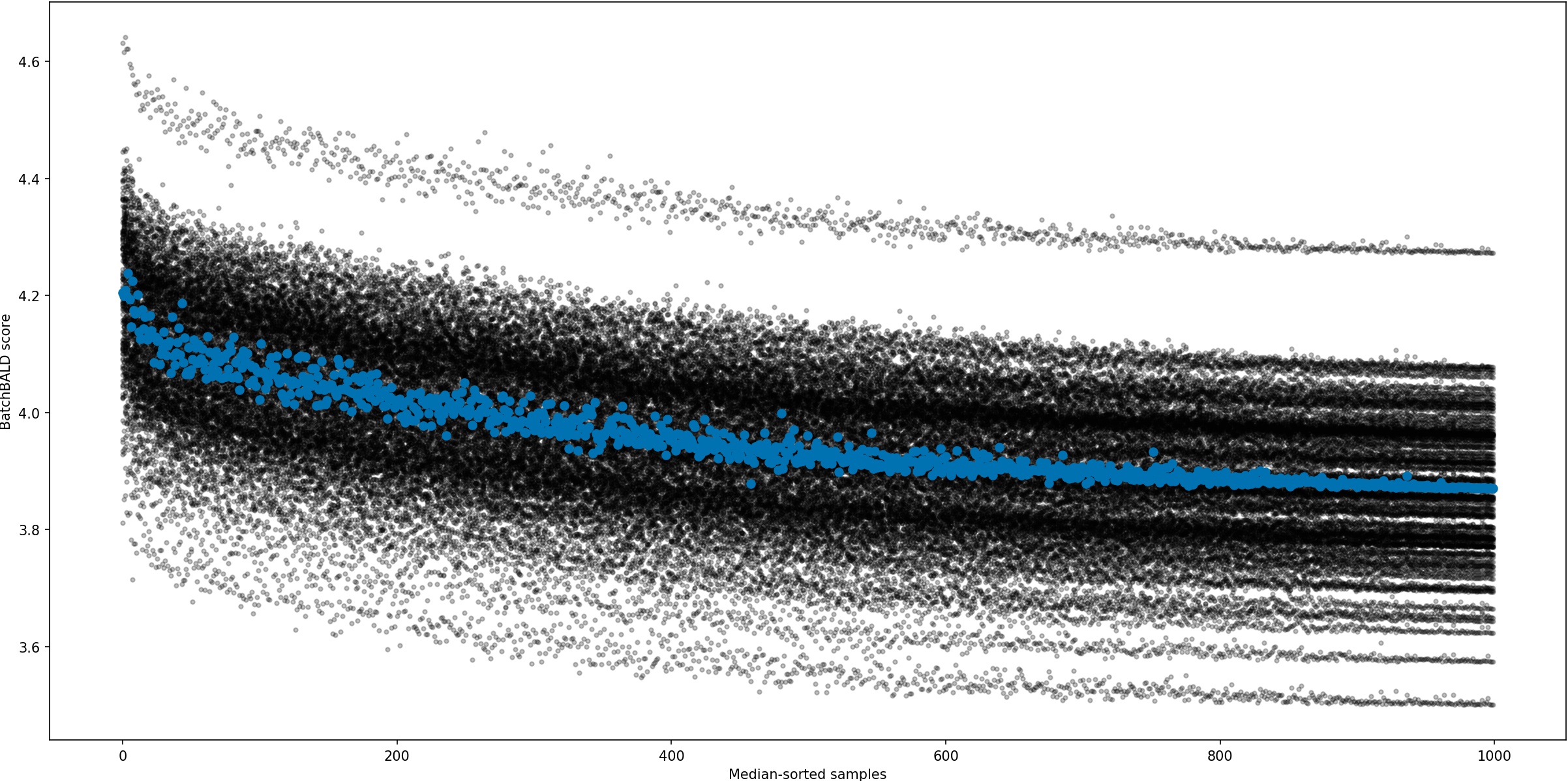
return joint_entropiesAcquisition Size Ablation
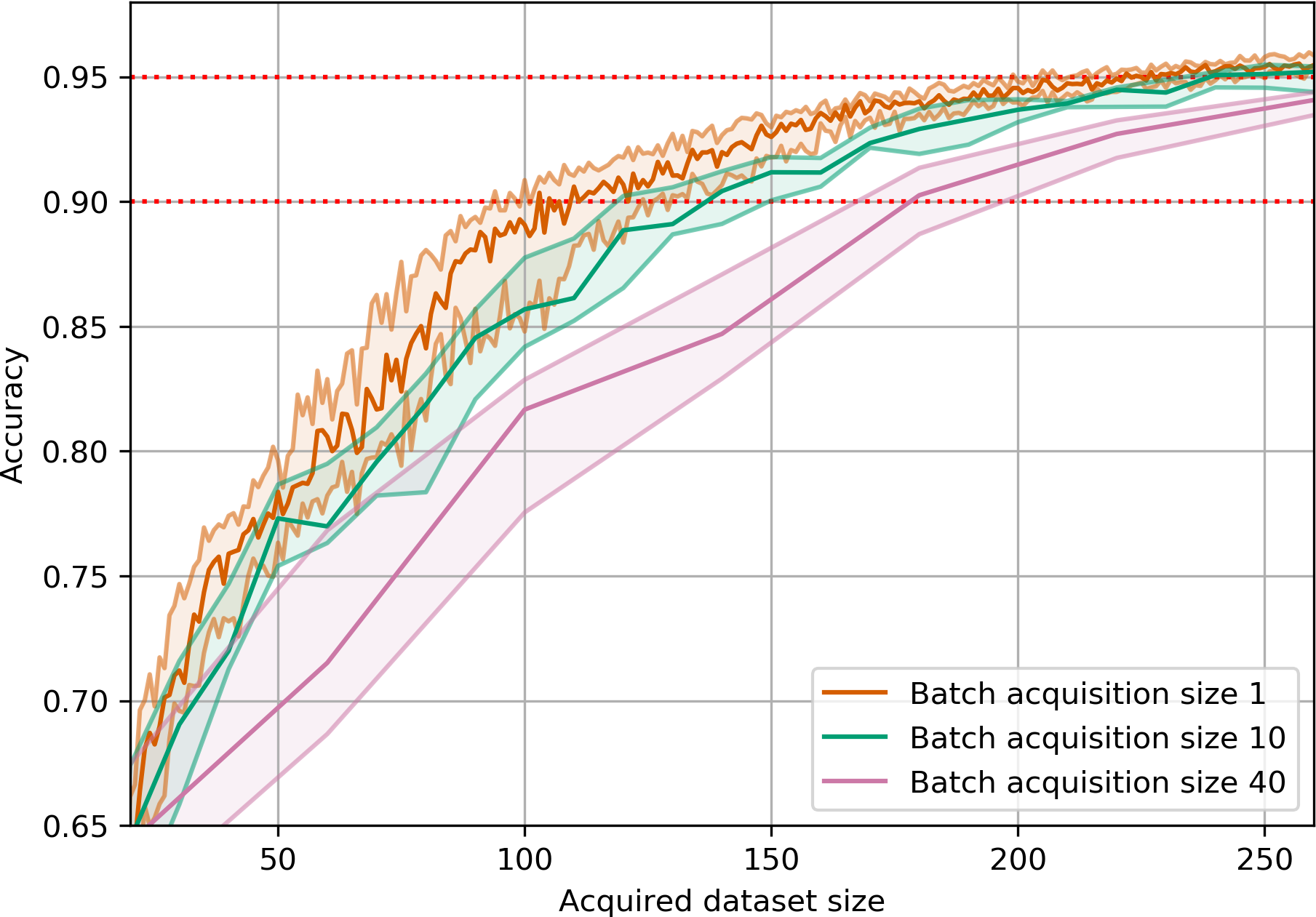
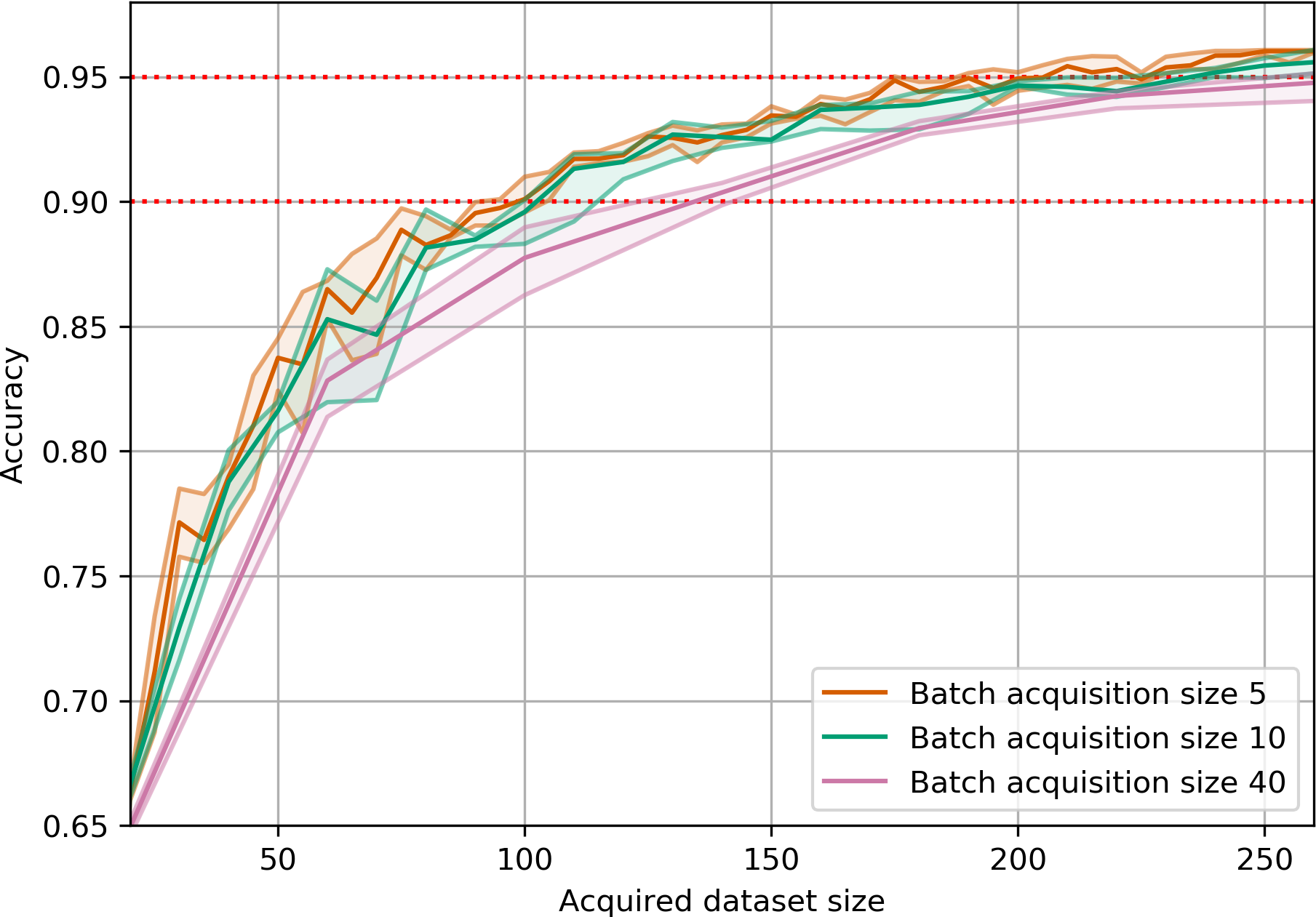
EMNIST
EMNIST is a dataset of 28x28 grayscale images of handwritten digits and letters.

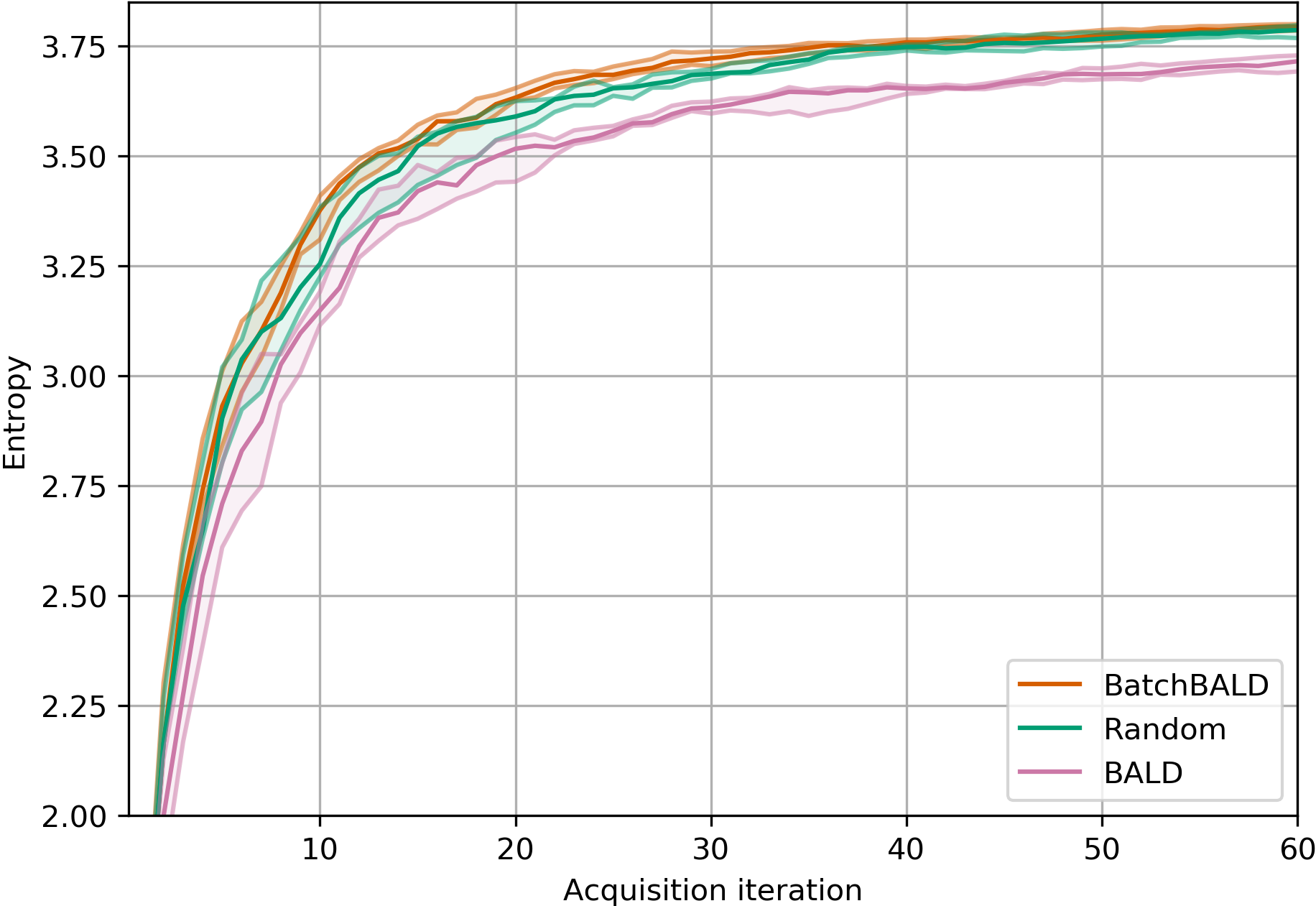
EMNIST Results
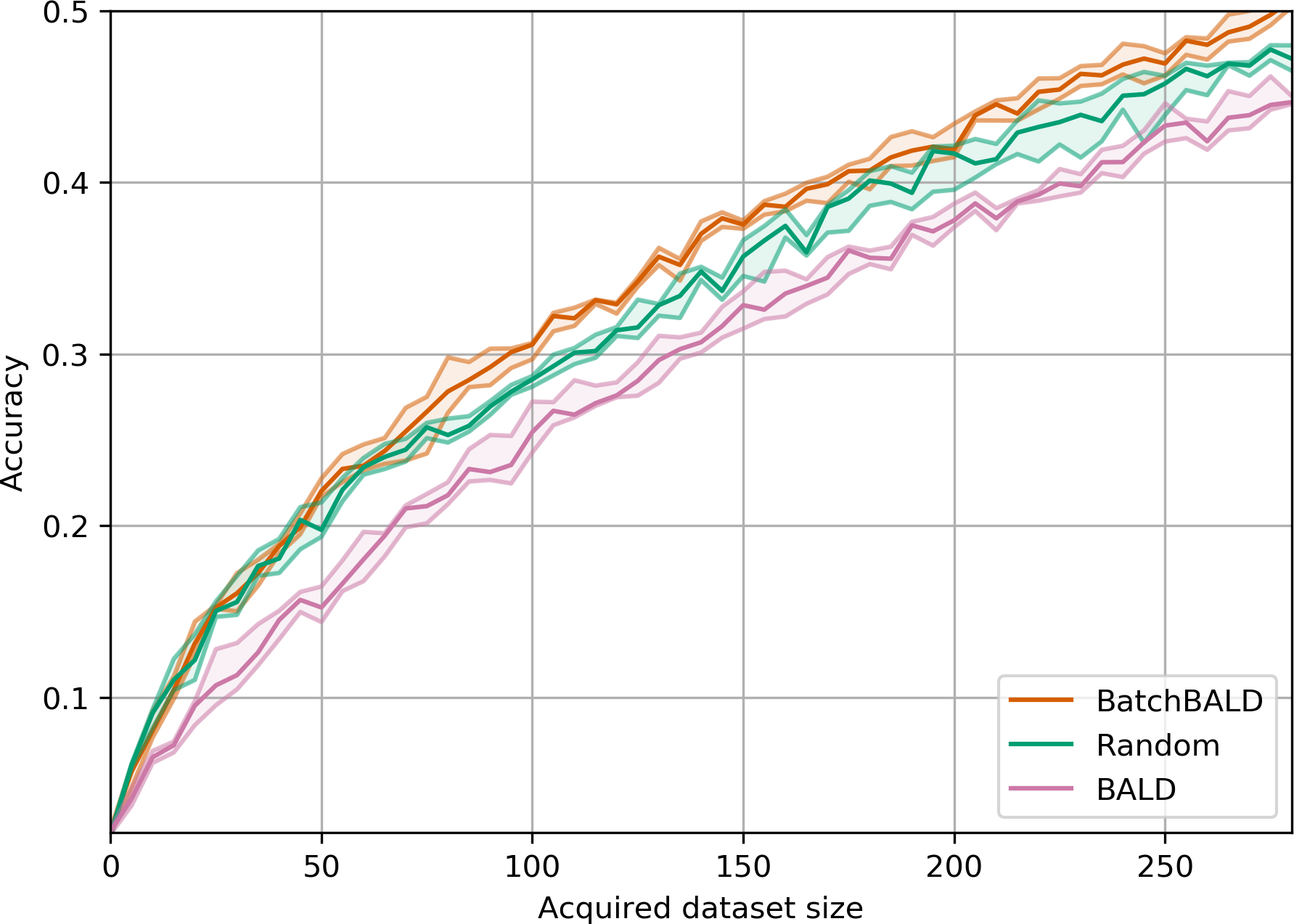
BatchBALD consistently outperforms both random acquisition and BALD while BALD is unable to beat random acquisition.
Acquired Label “Entropy”
Summary
- Naive batch acquisition can select redundant points
- BatchBALD uses joint mutual information
- Submodularity enables efficient subset selection
- Practical implementation considerations matter
Stochastic Batch Acquisition
The Problem
Traditional batch acquisition methods can be computationally expensive (in seconds)
K Top-K BADGE BatchBALD 10 0.2 ± 0.0 9.2 ± 0.3 566.0 ± 17.4 100 0.2 ± 0.0 82.1 ± 2.5 5,363.6 ± 95.4 500 0.2 ± 0.0 409.3 ± 3.7 29,984.1 ± 598.7 BatchBALD scales poorly with batch size (limited to 5-10 points)
Top-k selection ignores interactions between points
Need a more efficient yet simple approach that maintains diversity
Key Insight
Acquisition scores change as new points are added to training set
For BatchBALD, the difference is:
\[ \begin{aligned} &\MIof{\Y ; \W \given \x} - \MIof{\Y ; \W \given \x, \Y_{\text{train}}, \x_{\text{train}}} \\ &=\MIof{\Y; \Y_{\text{train}}; \W \given \x, \x_{\text{train}}} \\ &=\MIof{\Y; \Y_{\text{train}} \given \x, \x_{\text{train}}} - \MIof{\Y; \Y_{\text{train}} \given \W, \x, \x_{\text{train}}} \\ &=\MIof{\Y; \Y_{\text{train}} \given \x, \x_{\text{train}}} \ge 0. \end{aligned} \]
But if we don’t want to compute this?
Single-point scores act as noisy proxies for future acquisition value
Instead of deterministic top-k selection, use stochastic sampling
Sample according to score-based probability distribution
Noise Addition vs Sampling
We can add noise to the scores and take the top-K.
OR: we can sample from the score-based distribution directly.
This “duality” is similar to the Gumbel-Softmax trick.
Gumbel-Top-k Theorem
Theorem 4 For scores \(s_i\), \(i \in \{1, \ldots, n\}\), batch size \(k \le n\), temperature parameter \(\beta > 0\), and independent Gumbel noise \(\epsilon_i \sim \text{Gumbel}(0,\beta^{-1})\):
\[ \arg \text{top}_k \{s_i + \epsilon_i\}_i \]
is equivalent to sampling \(k\) items without replacement:
\[ \text{Categorical}\left(\frac{\exp(\beta \, s_i)}{\sum_j \exp(\beta \, s_j)}, i \in \{1, \ldots, n\}\right) \]
See also Kool, Hoof, and Welling (2019);Maddison, Tarlow, and Minka (2014); Gumbel (1954).
Key Implications
- Provides theoretical justification for stochastic batch acquisition
- Temperature \(\beta\) controls exploration-exploitation trade-off:
- \(\beta \to \infty\): deterministic top-k
- \(\beta \to 0\): uniform random sampling
Tip
Origin: The Gumbel-Softmax trick allows us to backpropagate through sampling operations, making it useful for both inference and training.
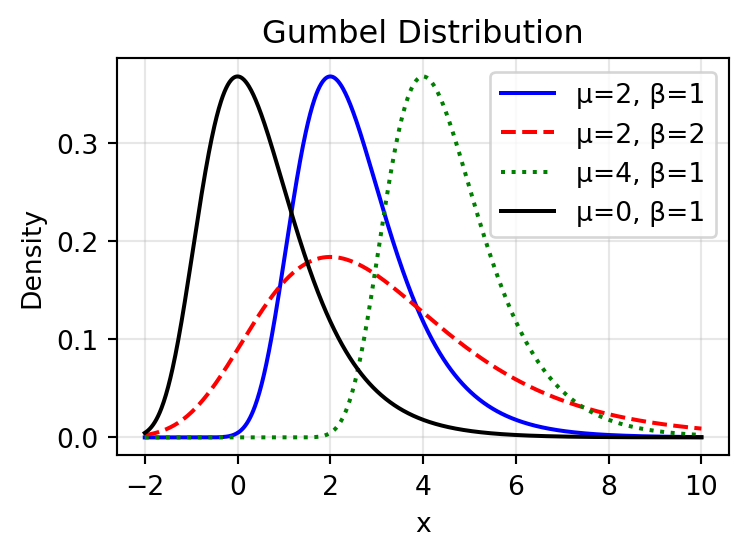
The Gumbel Distribution
The Gumbel distribution models the maximum of many random variables:
\[ F(x; \mu, \beta) = \exp\left(-\exp\left(-\frac{x-\mu}{\beta}\right)\right) \]
- \(\mu\) is the location parameter
- \(\beta\) is the scale parameter
Why Add Noise to Batch Selection?
The Problem with Top-K Selection
Assumption: Top-K assumes point scores are independent
Reality: Adding one point changes scores of all others
Key Insight: Most informative points cause biggest changes
Think About It…
What We Do:
- Score all points
- Pick top K at once
What Really Happens:
- Score points
- Add one point
- Model updates
- Scores change
- Repeat…
Score Correlation Over Time
Note
- Scores become less correlated as we add points
- Effect strongest for highest-scoring points
- Larger batches = more uncertainty
Details
Solution: Model the Uncertainty
- Add Noise: Acknowledge score uncertainty
- Sample Stochastically: Instead of deterministic top-K
- Benefits:
- Natural diversity
- Matches reality better
- Simple & efficient
How It Works
import numpy as np
from scipy.special import softmax
def stochastic_batch_acquisition(pool_set, batch_size, model, temperature=1.0):
# Score all points
scores = []
for x in pool_set:
score = acquisition_function(x, model)
scores.append(score)
# Convert to probabilities
probs = softmax(np.log(scores) / temperature)
# Sample without replacement
indices = np.random.choice(len(pool_set), size=batch_size, replace=False, p=probs)
return indicesBenefits
- Simple to implement (\(\mathcal{O}(|P| + K \log |P|))\) complexity) for Gumbel noise w/ top-K
- Naturally introduces diversity through stochastic sampling
- Performs as well as or better than sophisticated methods
- No hyperparameters to tune beyond temperature
- Compatible with any acquisition scoring function
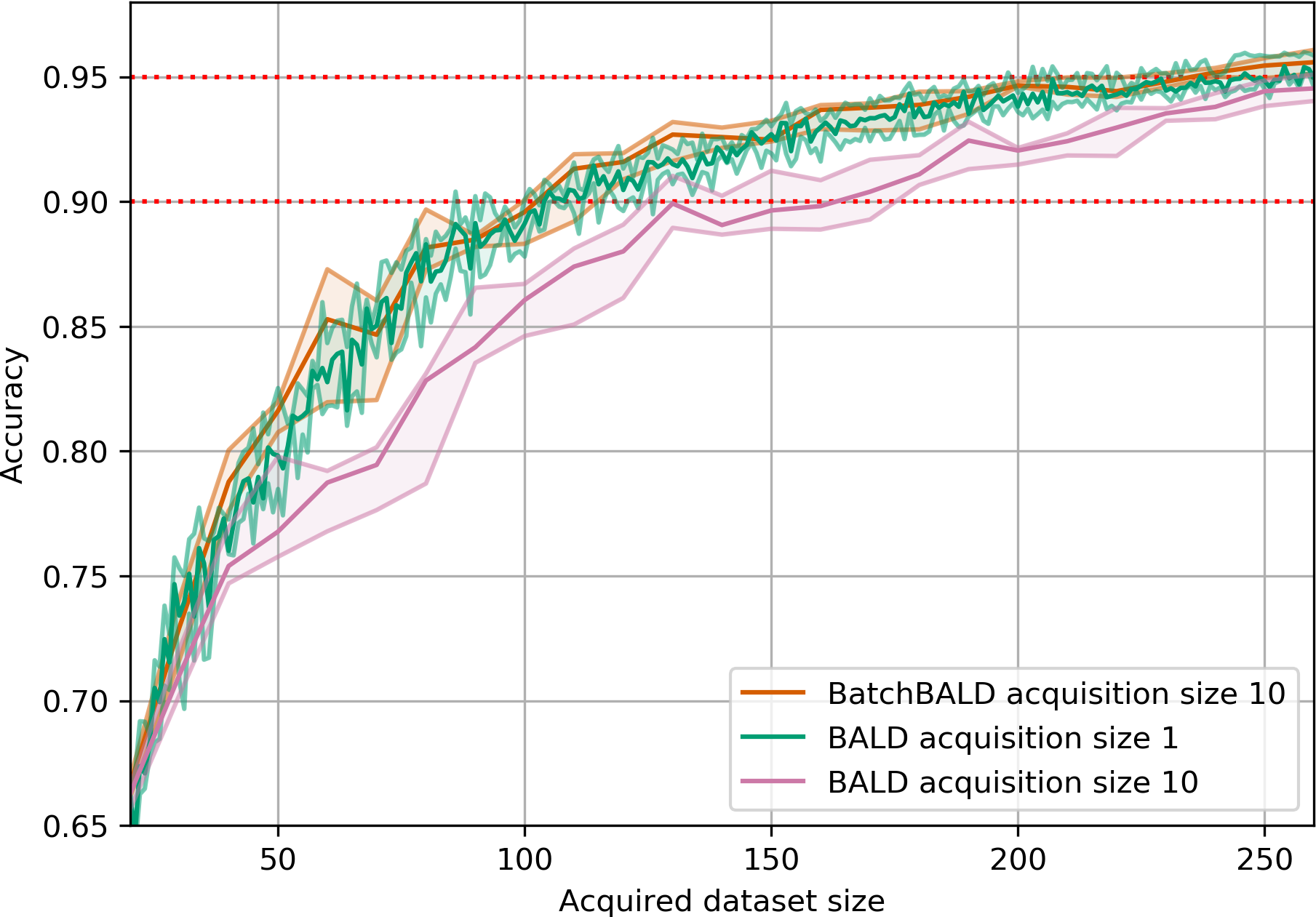
Results
Matches or outperforms BatchBALD and BADGE:
Repeated MNIST
EMNIST & MIO-TCD
Orders of magnitude faster computation
K Top-K Stochastic BADGE BatchBALD 10 0.2 ± 0.0 0.2 ± 0.0 9.2 ± 0.3 566.0 ± 17.4 100 0.2 ± 0.0 0.2 ± 0.0 82.1 ± 2.5 5,363.6 ± 95.4 500 0.2 ± 0.0 0.2 ± 0.0 409.3 ± 3.7 29,984.1 ± 598.7 Maintains performance with larger batch sizes
Avoids redundant selections common in top-k
Research Questions?
The strong performance of simple stochastic approaches raises serious questions about current complex batch acquisition methods:
- Do they truly model point interactions effectively?
- Are the underlying acquisition scores themselves reliable?
- Is the additional computational complexity justified given similar performance?
\(\implies\) Future work must develop more efficient methods that better capture batch dynamics while being tractable.